Introduction
PESI is Europe’s e-infrastructure for taxonomic information on species occurring in Europe.
The portal contains nearly 450,000 scientific names, 240,000 valid (sub)species names, and 140,000 vernacular names in 89 languages (see statistics).
The portal provides an integrated view of the major European checklists: Fauna Europaea (FaEu) for all European land and freshwater animals, Euro+Med Plantbase (E+M) with the vascular plants of Europe and the Mediterranean region, the European Register of Marine Species (ERMS) for species of the sea and coasts and the EU component of Index Fungorum (IF) for fungi. Still operating separately (different host institutions, committees, experts and servers) the register’s data is merged yearly in the PESI Data Warehouse and is available through this single portal.
In addition to taxonomic information, PESI harvests other information on species (images, literature, distributions, conservation status) and provides links to other portals (e.g. National check-lists, red species lists and other bioinformatics databases such as the Biodiversity Heritage Library for literature and the DNA databases of the Barcode of Life and GenBank).
The search interface is the main public access point to information on species living in Europe. However, the portal also provides services for those building their own species applications.
How to search information on the PESI portal?
The advanced search interface provides a number of fields to generate output based on selected parameters. This can be seen as applying filters on a database. The parameters are combinable, and none of the fields are mandatory, which means that a search without setting a parameter will output the entire database.
Taxonomic parameters
- (part of) ScientificName, Common name, Authority
- Equals/above/below taxon rank (e.g. species, family, class, …)
- Belonging to a group (a higher rank e.g. Mollusca)
Additional parameters
- Priority lists + Priority status (e.g. IUCN-endangered, Bird directive-protected, HYPPZ, ...)
- Occurrence + Occurrence status (Absent, Present, Introduced, ...)
The occurrence status “present” includes all other statuses except for “absent”. The list of areas is linked to the MarineRegions gazetteer (https://www.marineregions.org). This hierarchical gazetteer of place names makes it possible to add relationships to areas. The users can for example generate a list of species from France and the system automatically includes the species that are recorded as present in Corsica.
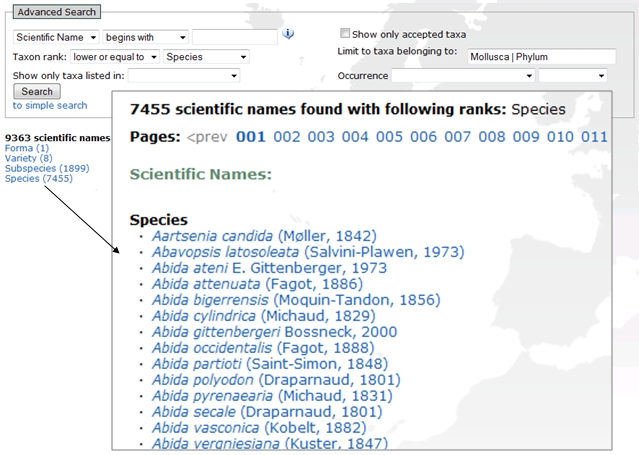
A combined screenshot which illustrates an example search and output of all species and infraspecies of mollusks.
Besides creating lists of species, the user can search for a particular taxon by entering (part of) the scientific name, name authority or common name. However, if there is no exact match, the search tool performs a number of ‘intelligent’ consecutive queries until matches are found:
- fuzzy match (Tony Rees’ TAXAMATCH algorithm), which checks on several spelling mistakes (more info: http://www.cmar.csiro.au/datacentre/taxamatch.htm).
- other potential genus-species combinations:
- FaEu model: it checks for reverse synonyms, e.g. if the species epithet occurs within genera that are synonym to the genus name you entered in the search box. For example, if you enter Avesaonchotheca blomei the portal will not find an exact match, but will suggest Aonchotheca caudinflata (Molin, 1858), because Avesaonchotheca is a generic synonym of Aonchotheca, and the species epithet blomei occurs in Capillaria blomei, which is a synonym of A. caudinflata.
- WoRMS model: checks if the species epithet occurs in other genera within the same Classis. For example, if you enter Parus merula, then the portal will not find an exact match, but will suggest Turdus merula, because it knows the genus Parus belongs to the Class Aves (=birds) and the species epithet merula occurs in the bird genus Turdus.
- checks if the name is present in the World Register of Marine Species (WoRMS)
- checks if the name is present in the Catalogue of Life (CoL)
- checks if the name is present in the Global Names Index (GNI).
Tools for taxonomic standardization
Taxon match, an ABC tool for species names
The correct spelling of a species name is not always trivial (which one is correct: Cirrhitichthys, Cirrhitychthys or Cirritichthys?) and it is very difficult for non‐taxonomists to keep up with the valid status of species names. PESI has developed a powerful online name matching tool to standardize your names with the PESI database. The tool returns standard PESI taxonomic information in a user-friendly format (e.g. MS Excel or tab-delimited text file). You need to upload a list of species names, match the columns with the fields in the PESI data warehouse and the system will return the file with valid names (notifies when the name is an unaccepted synonym), the authority and publication date, the hierarchical classification, quality status (expert validated or not) and the check-list’s Globally Unique Identifiers. When there are multiple matches the system provides a pick‐list. To avoid matching with homonyms, you can limit the query to a specific higher rank (e.g. Aves, Mollusca, etc). If your species list is restricted to a particular area, you can also check if this corresponds to the occurrences in the PESI database (via the “limit taxa belonging to a particular country” box). The tool is an implementation of the fuzzy matching algorithm written by Tony Rees (CSIRO, Australia), which comprises a suite of custom filters and tests used in succession on genus, species epithet, plus authority where supplied. We also used the Scientific Names Parser written by Dmitry Mozzherin. For more information on the taxon match algorithm, visit http://www.cmar.csiro.au/datacentre/taxamatch.htm.
In contrast to the taxon match, where the user has to upload a species list, the portal also provides a platform-independent SOAP/WSDL web service. This web service allows users to dynamically link their own applications to the PESI database and will allow them to match a locally stored species list and add taxonomic and additional information derived from PESI.
A few examples of possible applications:
- getGUID: Get the first exact matching GUID for a given name
- getPESIRecords: Get one or more matching (max. 50) PESIRecords for a given name
- getPESINameByGUID: Get the correct name for a given GUID
- getPESIRecordByGUID: Get the complete PESI Record for a given GUID
Globally Unique Identifiers (GUIDs)
Over the last couple of years the Biodiversity Informatics community underwent a vivid debate on the implementation of persistent Globally Unique Identifiers (GUIDs) for the object types to be networked in the emerging biodiversity data infrastructure (see http://wiki.tdwg.org/GUID). For PESI, the assignment of GUIDs is restricted to scientific names (i.e. nomenclatural entities and not name strings) and taxa (a scientific name used in a certain context). PESI will primarily be used as an authoritative resource for these two core object types and hence should offer persistent GUIDs to serve other infrastructures and networks.
Who will create GUIDs in the PESI Network?
Each participating checklist is responsible for assigning GUIDs to their objects. The GUIDs can be "raw" and do not have to follow a specific protocol (example: B85E62C3‐DC56-40C0-852A-49F759AC68FB, used by E+M) or can be based on the checklist's internal identifier systems and have the format of Life Science Identifiers (LSIDs). ERMS and FaEu have implemented LSIDs for all its taxonomic names. For example, the LSID for Solea solea is: urn:lsid:marinespecies.org:taxname:127160
You can resolve an LSID via the various services available. Example for Solea solea: http://lsid.tdwg.org
The returned model is RDF (XML) with metadata elements from Darwin Core and Dublin Core.
The checklist GUIDs are listed on every taxon page, and is part of the URL. You can use the search tool, the taxon match tool or the web service to add these GUIDs to your list of names.
Contribute data to PESI
The taxonomic information comes from four European checklists (ERMS, FaEu, E+M and IF). If you wish to contribute taxonomic information, you should contact the managers of these checklists.
If you have comments/remarks on a particular taxon, you can use the online feedback form. We will forward your message to the responsible person.
The PESI portal can establish links with other (national) species portals and exchange data and information in an automated
manner. There are currently a few successful cases:
1. Distribution data from the Atlas Florae Europaeae (AFE).
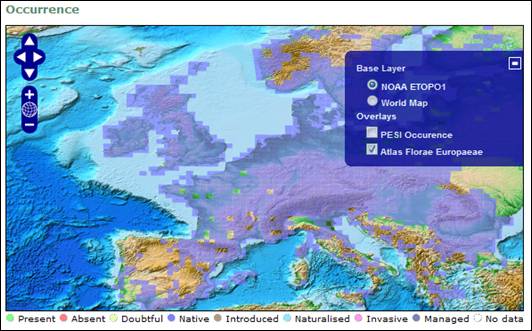
Atlas Florae Europaeae (AFE) provides its occurrence data via a Web Mapping Service (WMS) server set up at the University of Helsinki. They have been extracted from the AFE volumes (http://www.luomus.fi/english/botany/afe/index.htm) and displayed as a grid colored by the PESI Occurrence Statuses. These occurrences are restricted to a number of vascular plants described in AFE.
Screenshot of the distribution map, showing the occurrences of Atriplex patula, as served by the Atlas
Florae Europaeae.
2. Images from the Dutch Species Register (NLSR).
We have established a dynamic connection with the Dutch Species Register (NLSR). The
NLSR portal serves the information of species images in an RDF XML format,
which we harvest and display on the portal via a POST/GET request. The species
pages show thumbnail images and the metadata of each image (author + source).
When clicking on the thumbnail you are directed to the full image URL on the
source website.
It works as follows: when displaying a PESI taxon page the PESI web server contacts the NLSR web service at http://www.nederlandsesoorten.nl/get?site=nlsr&view=nlsr&id=i000091&action=search&searchString=Taxon. It sends one input parameter (searchString) which corresponds to the name of the taxon. The web service at NLSR responds with XML-structured data. This XML tells us whether there are images available for this particular taxon or not. If any references to images are present in the XML results, they contain absolute links to the image files at NLSR. When displaying the PESI taxon page these pictures are included in the HTML output by using the image URL's we retrieved from the web service.
For example:
- NLSR: http://www.nederlandsesoorten.nl/get?site=nlsr&view=nlsr&id=i000091&action=search&searchString=passer montanus
- PESI: http://www.eu-nomen.eu/portal/taxon.php?GUID=urn:lsid:faunaeur.org:taxname:97441
Screenshot of the thumbnail images served by the Nederlands Soorten Register.

Deep links to other biodiversity information systems
The PESI portal provides links to other portals (e.g. National check-lists, red species lists and other bioinformatics databases such as the Biodiversity Heritage Library for literature and the DNA databases of the Barcode of Life and GenBank).
1. The biodiversity heritage library (BHL) website is queried at http://www.biodiversitylibrary.org/services/pagesummaryservice.ashx?op=PageNameSearchForTitles&name=taxon name. The PESI web portal performs a regular expression match on the keys 'TitleCount' and 'PageCount'. When matches are found, the information is cached in our database for one month. The next time someone loads a particular taxon page within the same month, the cached information is displayed. If the request time compared to the last update in the database is above one month, the information is refreshed by re-querying the BHL web service.
2. GenBank provides a standard method for linking to their pages. We retrieve the information from http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?name=taxon name. When the link exists, it is displayed on the species page.
3. The Barcode of Life datbase (BOLD) results are fetched from http://www.boldsystems.org/views/taxbrowser.php?taxon=taxon name. Only when the 'specimens with barcodes' parameter is set, a link is visible on the species page.
Disclaimer
General disclaimer
The PESI Data Administrators reserves their exclusive right in its sole discretion to alter, limit or discontinue the Site or any Materials in any respect. The PESI Data Administrators shall have no obligation to take the needs of any User into consideration in connection therewith. The PESI Data Administrators reserves the right to deny in their sole discretion any user access to this Site or any portion thereof without notice. No waiver by the Data Administrators of any provision of these Terms and Conditions shall be binding except as set forth in writing and signed by its duly authorized representative.
No Warranty
PESI and its data suppliers provide this database free of charge for the benefit of the public in an “as is” condition. Neither PESI nor its data suppliers warrants, guarantees, or makes any representation regarding the accuracy, completeness, correctness, reliability, currency or otherwise, of the databases or the use or results to be obtained from using the databases or the information contained therein, or any related documentation or written materials. Neither PESI nor its data suppliers makes any representations or warranties whatsoever, express or implied, with respect to the database, and, in particular, PESI and its data suppliers disclaim all implied warranties including without limitation any warranties of merchantability, non-interference, non-infringement, informational content, or fitness for a particular purpose with regard to the database.
Limitations on liability
Except to the extent required by applicable law, in no event will PESI or its data suppliers be liable to you on any legal theory for any special, incidental, consequential, punitive or exemplary damages arising out of this license or the use of the work, even if licensor has been advised of the possibility of such damages.
Governance and Copyright
The content of the PESI databases is managed by the component databases, please refer to them individually.
The PESI portal will have a common approach to citation and Creative Commons licensing for all databases, including databases outside the SMBED committee. The copyright used will follow the Attribution-Share Alike scheme (for more information, view http://creativecommons.org/licenses/bysa/3.0/). Ideally, all data providers should abide by the same license requirements to avoid conflicting policy interactions if combined datasets from different sources are downloaded through the PESI facilities.
Terms of Use
By downloading or consulting data from this website, the visitor acknowledges that he/she agrees with the PESI data policy, and agrees to the following:
- If data is extracted from the PESI website for secondary analysis resulting in a publication, the PESI website should be cited following the citation scheme given below.
- Users must acknowledge the contribution of the relevant Data Providers in any derived information product or publication, whether printed, electronic or broadcast, that is based wholly or in part on the material, data and/or information they make available to them. Where users make specific use of or reference to a particular biological dataset they must acknowledge the original data provider (where their name is made available) using the referencing format provided within the PESI database.
- The citation system proposed for PESI generally follows that used in citing works published in hardcopy and so should be straightforward for researchers to use.
- [Author/s][Year]. [Chapter] In [Book]. [Publisher]. [Page]
- The template to be used for PESI will be as follows when verified by expert/contributed by European Species Database (ESD):
- Editors (Year). Taxon + Authority. In: ESD title or other Database or data manager. Accessed through the PESI at [URL] on [date].
- For example: Reid, David G.; Gofas, S. (2011). Littorina littorea (Linnaeus, 1758). Via: Costello, M.J.; Bouchet, P.; Boxshall, G.; Arvantidis, C.; Appeltans, W. (2011) European Register of Marine Species, Accessed through PESI at https://www.eu-nomen.eu/portal/taxon.php?GUID=urn:lsid:marinespecies.org:taxname:140262 on 2026-02-28
- Where the record has not been verified the database is the author, in which case the following template is used.
- PESI (Year). Taxon + Authority. Accessed through PESI at [URL] on [date].
- For example: PESI (2010). Salmo salar Linnaeus, 1758. Accessed through PESI at http://www.eu-nomen.eu/portal/taxon.php?GUID=urn:lsid:marinespecies.org:taxname:127186 on 2026-02-28.
To cite the entire database, use the following citation:
PESI (2026). Pan-European Species directories Infrastructure. Accessed through www.eu-nomen.eu/portal, at 2026-02-28
Citations (Google scholar)
Archive
The PESI portal only shows the latest version of the PESI datawarehouse. Previous versions are archived in Microsoft SQL server 2008 format and are stored at the VLIZ Marine Data Archive (MDA; https://mda.vliz.be). VLIZ is an official national data centre and the data on these servers are also stored on back-up servers and on tapes that are physically stored on a remote location.
Version history
- 2015-06-15 (v2.3): Second update of all component databases
- 2013-03-04 (v2.0): First update of all component databases
- 2011-04-18 (v1.0): Portal launch

